What is comprehensive genomic profiling for solid tumors?
Comprehensive genomic profiling (CGP) is an approach that relies on targeted next generation sequencing (NGS) to detect cancer biomarkers in a large number of genes. The precise definition of CGP is not currently regulated and therefore does not have a universally accepted classification. Though some may think of CGP as an approach that relies on a single assay, solutions to provide comprehensive tumor profiling can be modular and flexible with multiple assays that can be sequenced and analyzed at the same time. Regardless of the assay method, the goal of CGP is always to gain an extensive view into the genomic alterations in a sample to characterize the cancer.
High-quality NGS CGP will provide insights into relevant cancer biomarkers and genomic signatures, including:
- Single nucleotide variants (SNVs)
- Insertions and deletions (indels)
- Copy number variants (CNVs)
- Gene fusions
- Gene expression levels
- Structural variants
- Microsatellite instability (MSI)
- Tumor mutational burden (TMB)
Why is comprehensive genomic profiling used for solid tumors?
Relative to other approaches that aim to identify cancer-associated variants, i.e., single gene assays or small gene panels, CGP provides a larger variety of information about a sample. By obtaining a complete genetic profile of a sample, scientists can confidently detect all the major genomic changes present in hundreds of genes thereby reducing the risk of missing important alterations. Besides the volume and types of genomic variants identified, the larger content in CGP is often used for the detection of genomic signatures such as MSI and TMB. MSI status can provide insights into understanding tumor progression, as it may be correlated with response to some molecular agents that regulate immune and tumor cell interactions [1]. TMB is different from MSI in that it provides a measure of the mutations in a solid tumor sample, it similarly can be correlated in response to molecular agents related to immune and tumor cell interactions [2]. Though TMB can be measured by whole exome sequencing (WES), CGP methods for TMB have been shown to have high correlation with WES, but with a smaller sequencing resource footprint.
Is the future of CGP flexibility for each lab?
Not all molecular labs currently use CGP due to a variety of barriers including one-size-fits-all assays, workflow constraints, and high resource requirements. For more labs to benefit from answers that CGP analysis provides for solid tumors, the technology used needs to be able to be adapted to their needs, including having more content flexibility, resource optimization, and scalability.
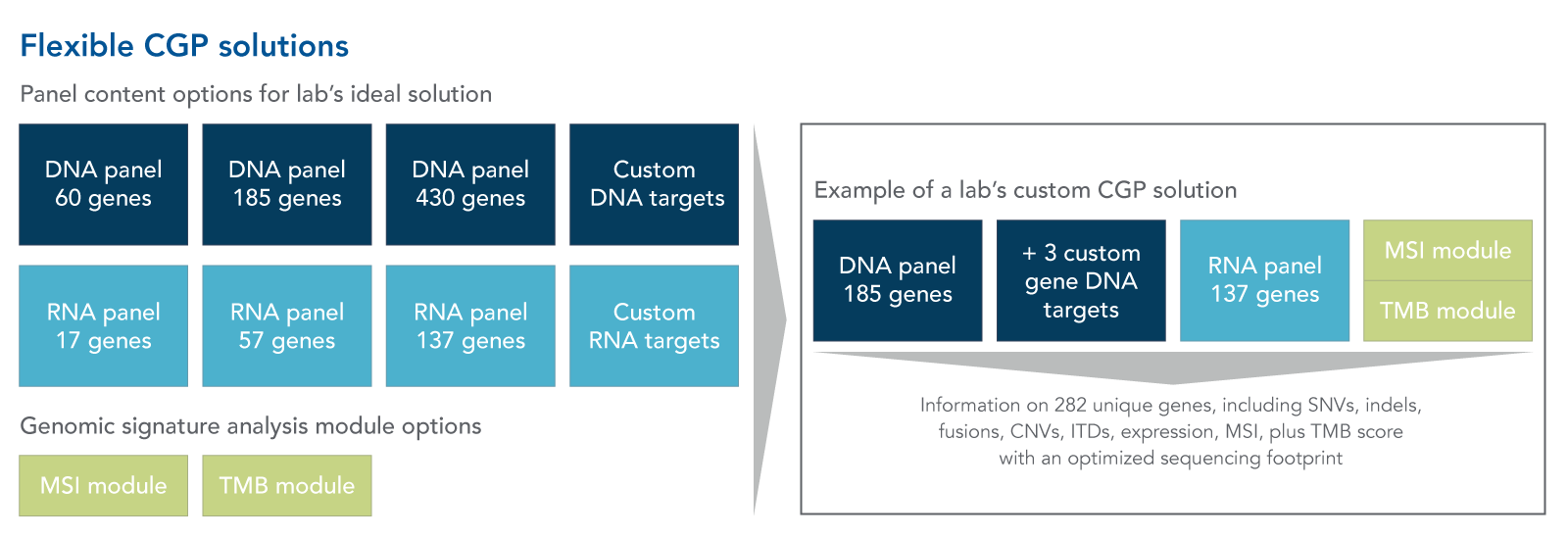
Content flexibility: As new cancer biomarkers are described, modular and custom CGP assays can offer the flexibility to rapidly add these targets to the panel as they are identified. The specific genomic alteration panel content and genomic signature analysis required can vary greatly between labs, so flexibility in content will be necessary for more labs to benefit from CGP analysis for solid tumors (Figure 1).
Resource optimization: With content flexibility comes the opportunity for CGP to be extremely efficient when it comes to minimizing the use of resources like samples, sequencing footprint, and computational power, without missing key information. The sequencing footprint of a custom CGP solution can be optimized because the reads obtained are only coming from the targeted genes. This both reduces the amount of noise in the sequencing reads and can save important computing resources by simplifying the downstream analysis of reads. Additionally, because CGP assays are designed to detect a wide variety of cancer biomarkers in a single sequencing run, the volume of sample needed as input can be reduced, relative to sequential biomarker assays. It may also be important for CGP assays to have workflows that can be automated as efficient use of time becomes increasingly important for labs.
Scalability: Finally, along with the flexibility and minimization of resources, liquid reagent formats for CGP assays enable high scalability with automation. CGP solutions can be designed for a lab’s current sample volume, with the ability to scale to a large number of samples sequenced in a single run.
CGP solutions with these benefits provide labs with a powerful ability to rapidly gather the insights needed to characterize cancers. These insights are vital to help scientists determine cancer progression and potential actionable targets in their precision medicine research.